Managing Pathology Research Data in an AI World
Share on social media
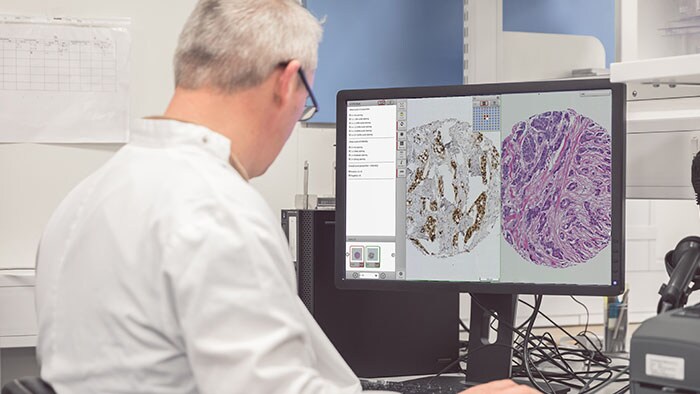
Tissue diagnostics and biomarker analytics are the keystones of cancer discovery research. However, delivering on the promise of personalized medicine requires multiple data sources to be integrated and analyzed. Management and analysis of large volumes of tissue samples are crucial to realizing the potential of personalized medicine. To accomplish this, imaging and pathology informatics support will be essential to moving forward in an increasingly complex and multifaceted medical research environment. There are several problems that a research platform needs to address if digital pathology is to prove an effective tool for drug and biomarker discovery studies.
Computational Pathology and AI – a rapidly growing disciple
The use of Artificial Intelligence (AI) and, in particular, Deep Learning in research is a growing area of interest for many. Management of pathology images is important in the development, validation and use of these algorithms, particularly within a research workflow. As with other disciplines, a large part of building AI algorithms involves curation of the images and annotations alongside any associated metadata. A recent publication by Campanella et al in Nature Medicine [1] used over 44,000 slides, showing the need to be able to manage datasets (images and associated data) of this magnitude.
When an algorithm is created, this data must be used along with metadata to test hypotheses. All of this means that ad-hoc methods of managing the high-resolution images inherent in digital pathology and their associated metadata and other derived data will inevitably cause difficulties in validating and reproducing large-scale studies. In this blog, we will attempt to highlight some of the challenges of such studies, and how they may be addressed through use of pathology image management solutions such as Xplore. [Not for diagnostic, monitoring or therapeutic purposes or in any other manner for regular medical practice]
Data management challenges in building and applying AI algorithms
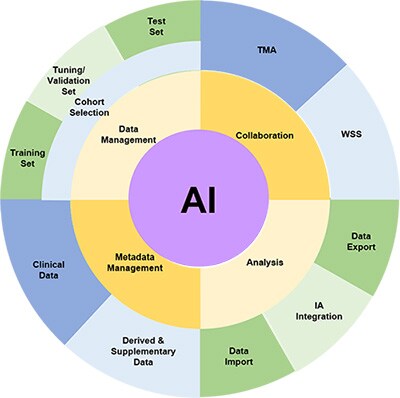
Cohort Selection
The fundamental step in identifying a biomarker is to select the cohort of slides which will be used for the study. This requires the ability to ingest digital images in various formats, store them and select those which are appropriate for a research study. There will also be metadata associated with such images such as clinico-pathological data, data source, antibody, etc. Associating this data with the images in the cohort is necessary, as misalignment will inevitably be a risk if these are held in separate databases or, as is often the case, spreadsheets and CSV files.
Splitting the image sets for good machine learning practice
It is generally understood within the Machine Learning community that it is good practice to split the available data into three subsets, and this is referenced in the recent FDA discussion paper, fda.gov/media/122535/download, as part of Good Machine Learning Practice (GMLP) – one which is used to train the algorithm(s), one which is not used within the training, and is used by the data scientists to validate and tune their model architecture and approaches, and one which is not used at all during the training and validation steps and is used solely to test the algorithm performance on completely unseen data. In order to ensure complete independence of the unseen test set, the ability to restrict permissions to view the test prevents inadvertent data ‘leakage’ from the test set to the training.
It is also necessary to ensure that the three sets of data (training, tuning and test) are drawn from the same distributions, so the ability to use metadata stored in the image management system to ensure this (in as much as it is possible) is a key feature.
Collaboration with Pathologists
[1] Campanella, Gabriele, et al. "Clinical-grade computational pathology using weakly supervised deep learning on whole slide images." Nature medicine 25.8 (2019): 1301-1309.
Very often, pathologists are intimately involved in the validation of algorithms and their scores and annotations are needed to provide the ‘ground truth’ against which the algorithm will be compared. The ability to collect pathologist scores and input and store these alongside the images is invaluable (both for WSIs and TMAs) and allows the data to be analysed alongside quantitative results obtained from the algorithms. This can not only be the correlation analyses and other measures of technical performance of the algorithms, but since the clinic-pathological data is also available for the images, the use of the algorithmic output can be compared to pathologist scores in subsequent survival/response analysis, paving the way for the use of algorithmic scores as an alternative to laborious and inconsistent manual pathology scoring for large-scale biomarker studies.